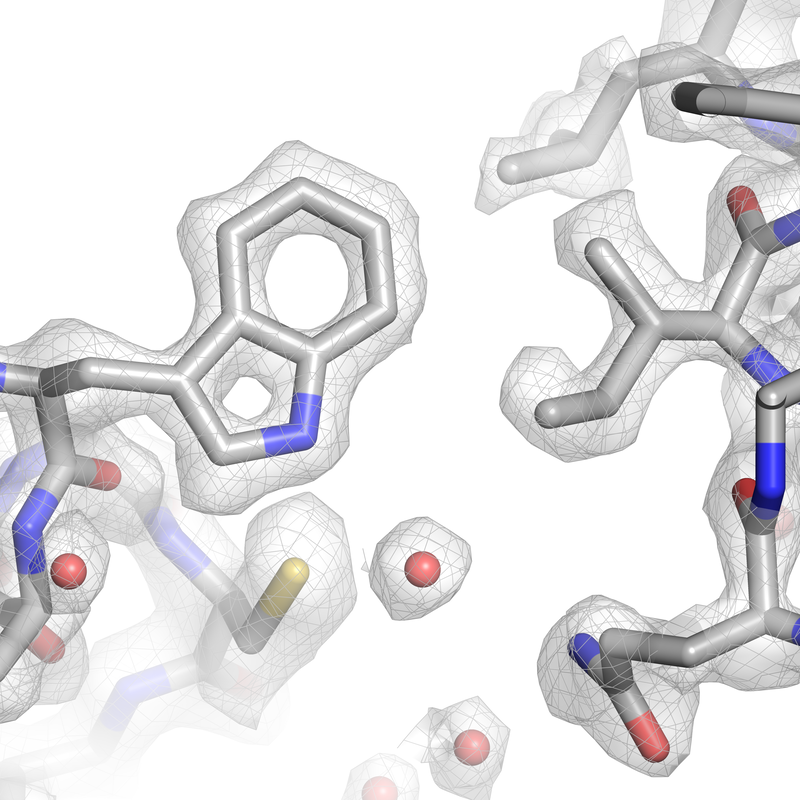
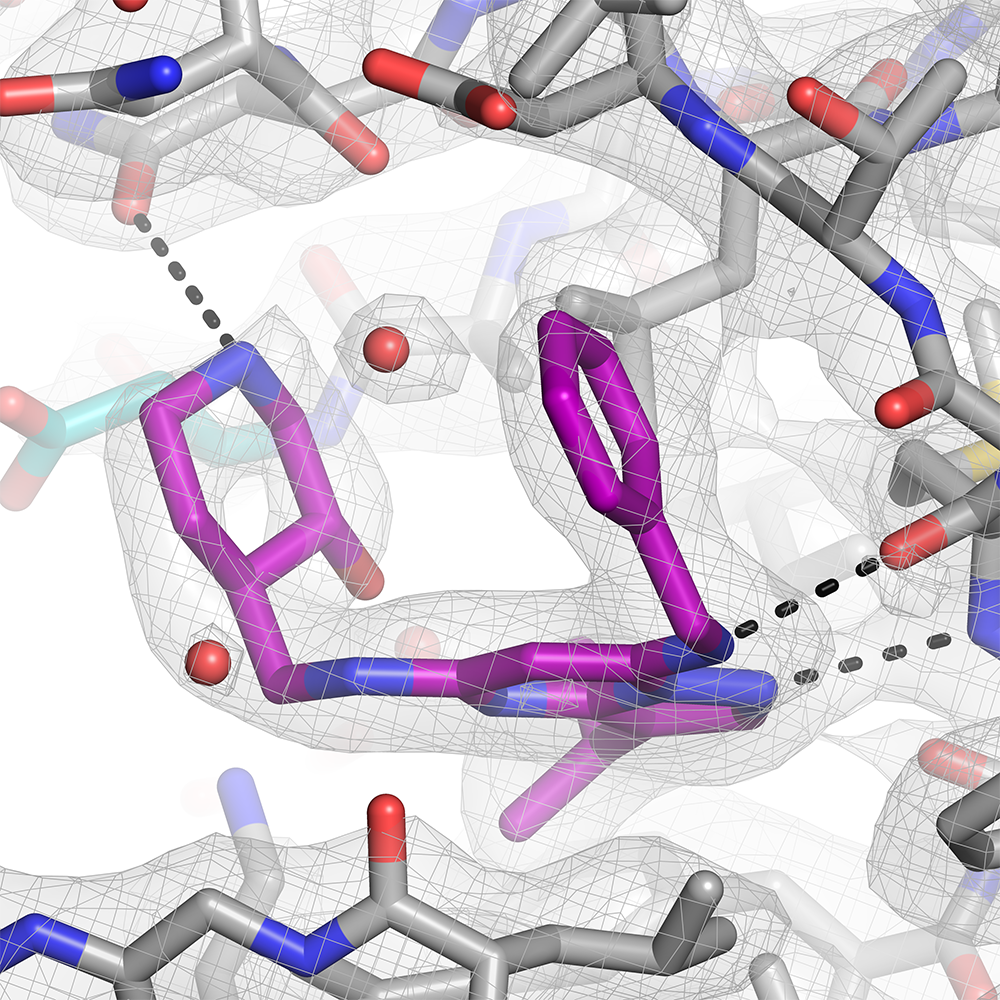
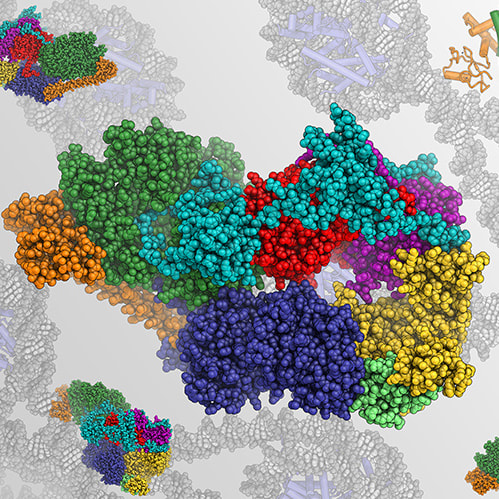
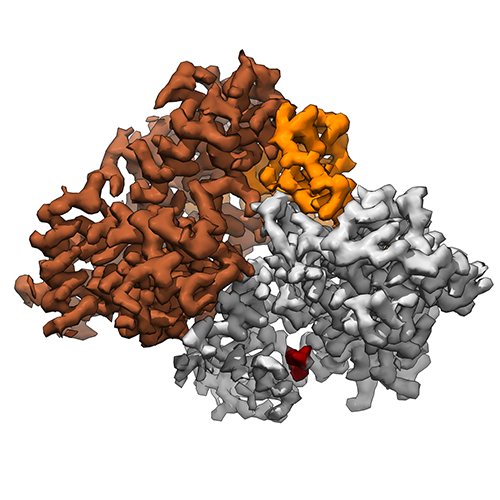
Learn more about the group
We are part of the Division of Structural Biology at the Institute of Cancer Research in London.
|
Research
Explore our research on nucleotide excision repair and read about challenges in cryo-EM
|
The Lab
|
Publications
|
News from the lab
Another new publication!March 13th, 2024
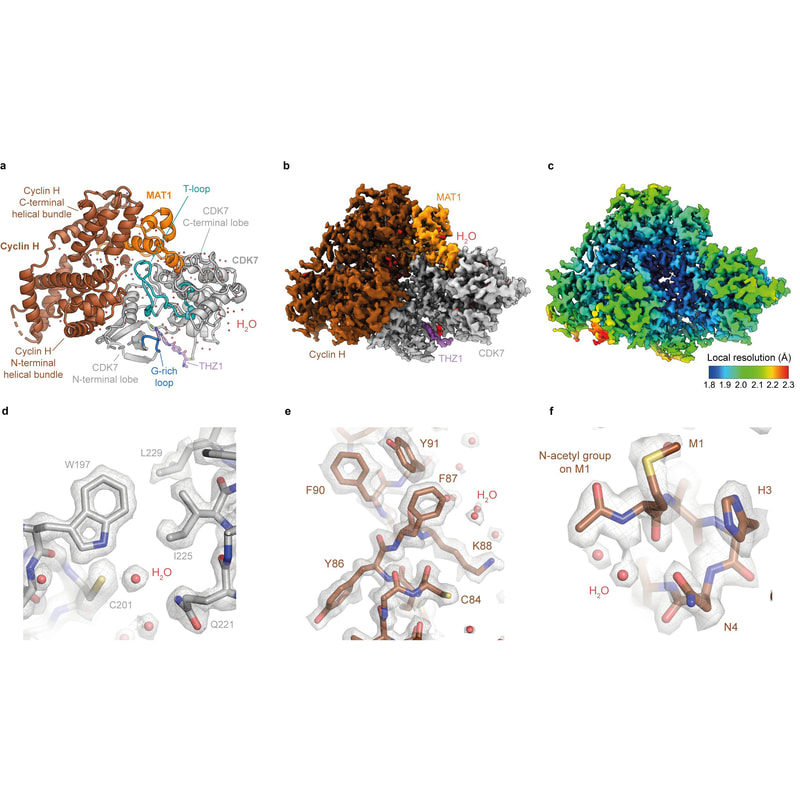
In collaboration with the groups of Prof. Simak Ali (Imperial College London) and Dr. Abhay Kotecha (Thermo Fisher Scientific), we established high-resolution/high-throughput cryo-EM workflows tailored for structure-based drug design. Applying these workflows to the ligand-bound human CDK-activating kinase, we determined more than a dozen structures at 2 Å resolution. Read all about it at Nature Communications. |
New publication!February 16th, 2024
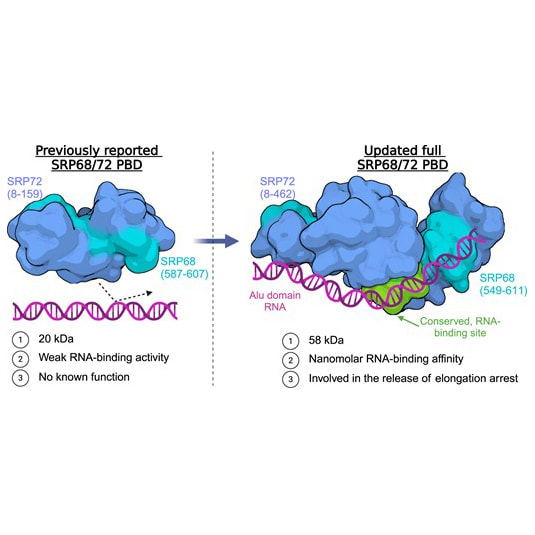
In collaboration with the groups of Prof. Sandro Ataide (University of Sydney) and Dr. Abhay Kotecha (Thermo Fisher Scientific), Junjie determined the structure of the protein-binding domain of the SRP68/72 dimer, which forms part of the eukaryotic signal recognition particle. Read all about it at Nucleic Acids Research. |
New job opening!May 6th, 2023
We are looking for a post-doctoral training fellow to join the lab and pursue structural and functional studies of DNA repair and related processes. Candidates with prior expertise in the biochemistry of nucleic acid-protein complexes or in structural biology/cryo-EM are encouraged to apply. Application deadline is June 4th, 2023. |
New pre-print!April 12th, 2023
We just published the lab's first pre-print! Together with collaborators at Thermo Fisher Scientific and Imperial College, we developed high-resolution and high-throughput cryo-EM workflows for structure-based drug design. We screened large numbers of grids and determined almost 20 structures of inhibitor-bound CAK at 2 Å from the best ones. Have a look on bioRxiv! |
New lab space & new lab membersAugust 23, 2021
We have moved into our newly refurbished lab space a few weeks ago (pictured on the left in its pristine state, though now already filled with lots of instruments and consumables). Junjie has joined the lab as a postdoc at the beginning of the month. Victoria (PhD student) and Natalia (postdoc) will join in the next few months to complete the team. |
We are (still) recruiting!June 1, 2021
Two new lab members have been recruited: Junjie and Victoria will join the lab in the summer and fall, respectively. To expand our team, we are still looking for an MRC-funded post-doctoral training fellow to join the lab. Candidates with prior expertise in the biochemistry of nucleic acid-protein complexes or in structural biology are encouraged to apply. Learn more about this opportunity here. |
Structure of the CDK-activating kinase bound to ICEC0942January 19, 2021
Our latest paper reports the 2.5 Å-resolution structure of the CDK-activating kinase bound to the inhibitor ICEC0942 and has been published in the Biophysical Journal. This work was a collaboration with the laboratory of Prof. Simak Ali at Imperial College London and the data were collected during Basil's final days in the Nogales Lab in Berkeley. |
Read about our workOctober 6, 2020
Céleste Nilges spoke to Basil about his past work and the new lab. Read about it in this blog post on the ICR website. |
Raw data available on EMPIAROctober, 2020
The raw data used for structure determination of the CDK-activating kinase is now available on EMPIAR: CAK-nucleotide complex: www.ebi.ac.uk/pdbe/emdb/empiar/entry/10431/ CAK-THZ1 complex: www.ebi.ac.uk/pdbe/emdb/empiar/entry/10438/ |
Structure of the human CDK-activating kinaseAugust 27, 2020
Basil's work on the structure of the CDK-activating kinase, performed in the Nogales Lab in Berkeley, has been published in PNAS. |