TFIIH: At the crossroads of eukaryotic transcription and DNA repair
The expression of the genetic information stored in the genome provides the protein and RNA molecules of the cell and is of pivotal importance for all living organisms. Gene expression starts with the transcription of the DNA sequence into RNA by RNA polymerases. More specifically, RNA polymerase II (Pol II) synthesizes the messenger RNAs (mRNAs) required for the production of proteins in all eukaryotic cells. Regulation of the amount of mRNA synthesized is an important and tightly controlled means of regulating the amount of protein production, and mis-regulation of transcription is associated with human disease, including cancer. In addition to Pol II, which performs the template-directed synthesis of the mRNA, multiple protein factors are required to support the various steps of transcription initiation. Transcription factor IIH (TFIIH) is a 10-subunit complex that functions as a general transcription factor for initiation by Pol II and is additionally crucial in nucleotide excision DNA repair. Mutations in the subunits of TFIIH lead to the severe inherited disease syndromes xeroderma pigmentosum, Cockayne syndrome, and trichothiodystrophy. The aim of Basil Greber's post-doctoral work in the laboratory of Prof. Eva Nogales at UC Berkeley was to determine the complete structure of TFIIH.
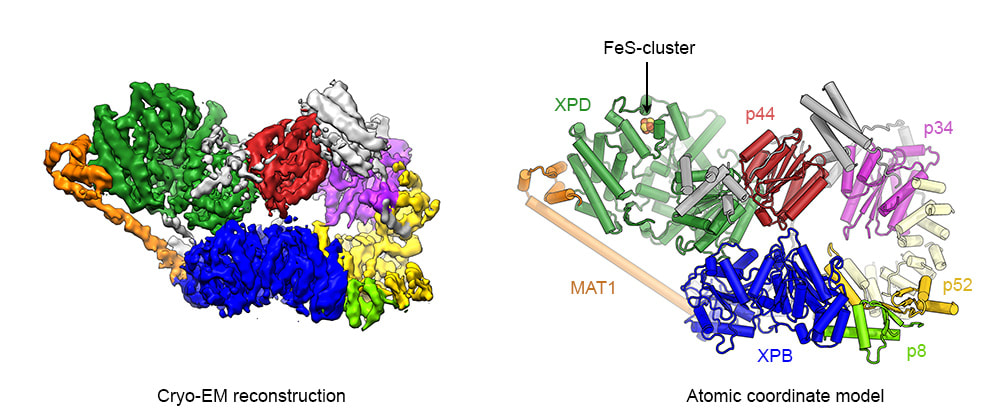
The cryo-EM structure of human TFIIH at 4.4 Å resolution
|
The cryo-EM structure of TFIIH at 4.4 Å resolution. Left: Cryo-EM map colored according to assigned subunits. Right: Refined atomic coordinate model of human TFIIH. Protein subunits are color coded and labeled.
Greber BJ, Nguyen THD, Fang J, Afonine PV, Adams PD, Nogales E (2017). The Cryo-Electron Microscopy Structure Human Transcription Factor IIH. Nature 549 (7672): 414-417. Epub 2017 Sep 13. PubMed |
TFIIH is composed of two subcomplexes, the TFIIH core complex and the CdK-activating kinase (CAK subcomplex). The core complex harbors the two DNA helicases/ATPases XPD and XPB and is sufficient for TFIIH to carry out its DNA repair function. For transcription initiation, the CDK-activating kinase (CAK) subcomplex, which harbors the kinase activity of CDK7 is additionally required. Using a Titan low-base cryo-electron microscope equipped with a side-entry holder and a K2 Summit direct electron detector, we initially determined the cryo-EM structure of human TFIIH at 4.4 Å resolution. This structure revealed the molecular architecture of the TFIIH core complex and the structures of the helicase cassettes of human XPD and XPB. Furthermore, we were able to map numerous human disease mutations in XPD and visualize the interaction of the CAK-subunit MAT1 with XPD and XPB. This interface is crucial for the dynamic recruitment of the CAK subcomlex to TFIIH and therefore for transcription initiation.
The complete structure of the human TFIIH core complex
In our previous structure, several important regions of the TFIIH core complex could not be fully interpreted due flexibility or limited resolution. Therefore, in an effort to improve the cryo-EM reconstruction and enable building of a complete model for the TFIIH core complex, we collected cryo-EM data using a Titan KRIOS microscope equipped with a Volta phase plate (VPP). The VPP enhances the low-resolution contrast of the images, which leads to better classification and alignment of the particle images. From these data, we obtained a cryo-EM map of human TFIIH at 3.7 Å resolution, which enabled building of a complete coordinate model of the TFIIH core complex. This new structure revealed the architecture of p62, the molecular mechanism of the recruitment of XPB to TFIIH, and suggests that XPD is a highly regulated enzyme. Several other TFIIH subunits perform XPD-regulatory roles, including blockage of functionally important interfaces, such as the ATP binding pocket and the substrate-binding cavity.
|
|
|
The cryo-EM structure of TFIIH at 3.7 Å resolution. Left: Movie that shows the structure of human TFIIH (protein subunits are colored: XPD green, XPB blue, p62 cyan, p52 gold, p44 red, p34 purple, p8 light green, MAT1 orange). Right: Conformational changes of human TFIIH during incorporation into the Pol II-PIC. The movie is based on the data from Greber et al. (2019), eLife 8: e44771 and He et al. (2016), Nature 533(7603): 359-365.
Greber BJ, Toso DB, Fang J, Nogales E (2019). The Complete Structure of the Human TFIIH Core Complex. eLife 8: e44771. Epub 2019 Mar 12. PubMed
Greber BJ, Toso DB, Fang J, Nogales E (2019). The Complete Structure of the Human TFIIH Core Complex. eLife 8: e44771. Epub 2019 Mar 12. PubMed
TFIIH is a dynamic molecular machine. During transcription initiation, it functions in the context of the pre-initiation complex (PIC), which contains Pol II, a number of additional general transcription factors, and promoter DNA. A comparison of our structure of free TFIIH with the structure of TFIIH in the context of the PIC (determined previously Dr. Yuan He and colleagues in the Nogales laboratory) reveals that TFIIH undergoes a conformational rearrangement upon entering the PIC and DNA binding. During this conformational change, XPB and XPD, which contact each other in free TFIIH, move apart. Additionally, a long helix of the MAT1 subunit connects XPB and XPD in free TFIIH. This interaction is no longer visualized in the maps of the Pol II-PIC. The transition from a closed to an open conformation of TFIIH may be linked to the engagement and translocation of DNA by XPB.
The structure of the human CDK-activating kinase
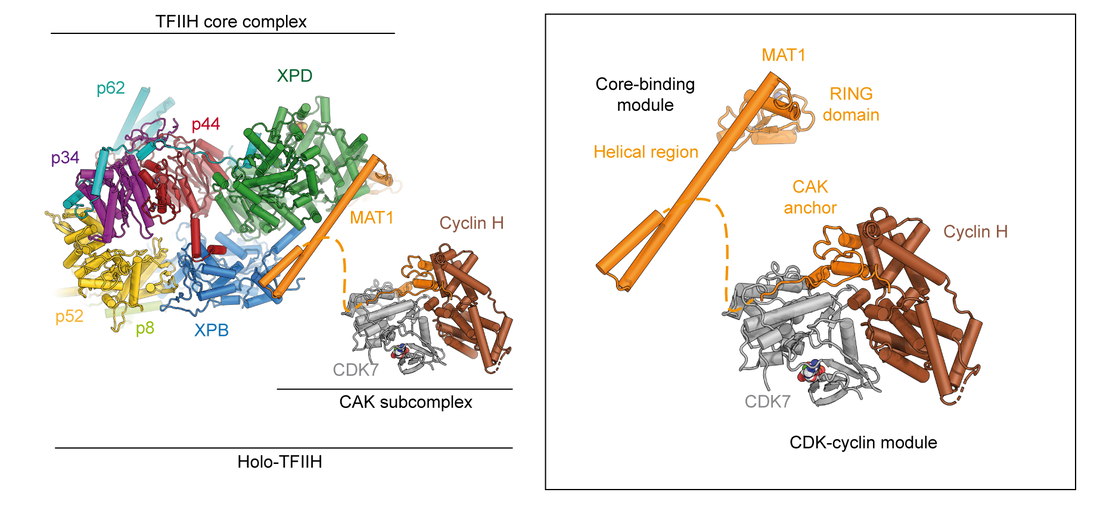
Due to its importance in transcription initiation, we have more recently also determined the structure of the CAK subcomplex (Fig. 1A) [6], revealing how the C-terminal domain of MAT1 interacts with CDK7 and cyclin H to stabilize the assembly. Additionally, MAT1 interacts with the CDK7 T-loop, thereby promoting an activated conformation of the kinase. In addition to its biological relevance, this structure at 2.8 Å resolution was also one of the smallest asymmetric complexes for which better than 3 Å-resolution had been achieved using cryo-EM at the time of publication.
|
The structure of the human CDK-activating kinase. Left: Structure of holo-TFIIH, composed of the core and CAK subcomplexes. Right: Structure of the human CAK, assembled from the structures of the isolated kinase module and the core-binding module resolved with the core complex.
Greber BJ#, Perez-Bertoldi J M, Lim K, Iavarone AT, Nogales E# (2020). The Cryoelectron Microscopy Structure of The Human CDK-Activating Kinase. Proc. Natl. Acad. Sci. U.S.A. 549 (7672): 414-417. doi: 10.1073/pnas.2009627117 Epub 2020 Aug 27. #corresponding authors PubMed |
For additional information on eukaryotic transcription, please visit the website of the laboratory of Prof. Eva Nogales at UC Berkeley.