High-resolution cryo-electron microscopy and its application to challenging targets
In order to enable biological discovery, we need to address challenges that arise during structure determination of biomolecular complexes by cryo-electron microscopy (cryo-EM). We are particularly interested in three challenges:
Structural flexibility: The molecular assemblies that we are working with are flexible. Some exhibit intrinsic continuous flexibility, others require the ability to undergo structural transitions to perform their biological function. We aim to exploit the ability of cryo-EM to visualise flexible assemblies and combine these results with biophysical methods to create "molecular movies" of the processes we study.
Fragile complexes: Interaction of molecular complexes with the air-water interface in the extremely thin film of sample that is generated during cryo-EM specimen preparation can lead their denaturation and destruction. We aim to exploit available techniques to circumvent these issues and obtain structures of intact and fully functional fragile molecular assemblies.
Structural flexibility: The molecular assemblies that we are working with are flexible. Some exhibit intrinsic continuous flexibility, others require the ability to undergo structural transitions to perform their biological function. We aim to exploit the ability of cryo-EM to visualise flexible assemblies and combine these results with biophysical methods to create "molecular movies" of the processes we study.
Fragile complexes: Interaction of molecular complexes with the air-water interface in the extremely thin film of sample that is generated during cryo-EM specimen preparation can lead their denaturation and destruction. We aim to exploit available techniques to circumvent these issues and obtain structures of intact and fully functional fragile molecular assemblies.
|
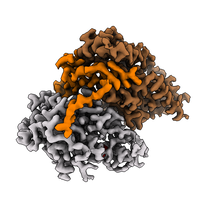
Cryo-EM map of the human CDK-activating kinase, with CDK7 shown in grey, cyclin H in brown, and MAT1 in orange. The map encompasses approximately 85 kDa of protein mass. At the time of publication, this was one of the smallest asymmetric protein complexes resolved to better than 3 Å by cryo-EM.
Greber BJ#, Perez-Bertoldi J M, Lim K, Iavarone AT, Nogales E# (2020). The Cryoelectron Microscopy Structure of The Human CDK-Activating Kinase. Proc. Natl. Acad. Sci. U.S.A. 549 (7672): 414-417. doi: 10.1073/pnas.2009627117. #corresponding authors PubMed |
High resolution structure determination of small complexes: Cryo-EM has the proven potential to achieve true atomic resolution for relatively large and rigid biomolecular complexes. However, many therapeutically relevant proteins or protein complexes are comparably small and more difficult to visualise at high resolution. At the same time, applications such as structure-based drug design benefit from high-resolution structural data. We are therefore interested in high-resolution structure determination of small complexes. Our initial work on the CDK-activating kinase was the first example of these efforts.
Cryo-EM for structure-based drug design
Due to its involvement in the regulation of transcription and the cell cycle, the human CDK-activating kinase is a promising target for cancer therapeutics, and several compounds have entered clinical trials. To enable structure-based discovery of next-generation therapeutics, we are working to improve the resolutions of our cryo-EM maps and determine structures in complex with clinical inhibitors.
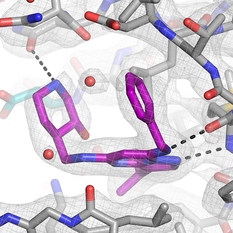
We initially determined the structure of the human CDK-activating kinase in complex with the selective inhibitor ICEC0942, a small-molecule compound that is currently undergoing clinical trials for cancer therapy. At an improved 2.5-Å resolution, we were able to accurately fit the inhibitor into the density, which revealed notable conformational differences compared to the structure of the same compound bound to CDK2. These results may provide avenues towards design of more specific CDK7 inhibitors.
|
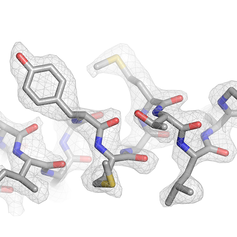
Cryo-EM map of the inhibitor ICEC0942 bound to the human CDK-activating kinase. The density allows the fitting of the substituents on the inhibitor and reveals the location of water molecules.
Greber BJ#, Remis J, Ali S, Nogales E (2021). 2.5 Å-resolution structure of the human CDK-activating kinase bound to the clinical inhibitor ICEC0942. Biophys. J. doi: 10.1016/j.bpj.2020.12.030 Epub 2021 Jan 19. #corresponding author |
Most recently, we were able to improve the resolution of our cryo-EM reconstructions of the human CDK-activating kinase to better than 2 Å by using a high-end microscope equipped with a cold field emission gun (cold-FEG) and a latest-generation energy filter. Based on this result, we established high-throughput and high-resolution cryo-EM workflows that allowed us to determine almost 20 inhibitor-bound structures of the human CDK-activating kinase at 1.8-2.1 Å resolution. Improving both the resolution and the throughput of cryo-EM is important for applicability of the method in structure-based drug design. This work was performed in collaboration with the groups of Dr. Abhay Kotecha (Thermo Fisher Scientific) and Prof. Simak Ali (Imperial College).
|
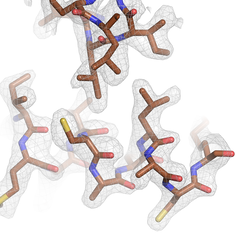
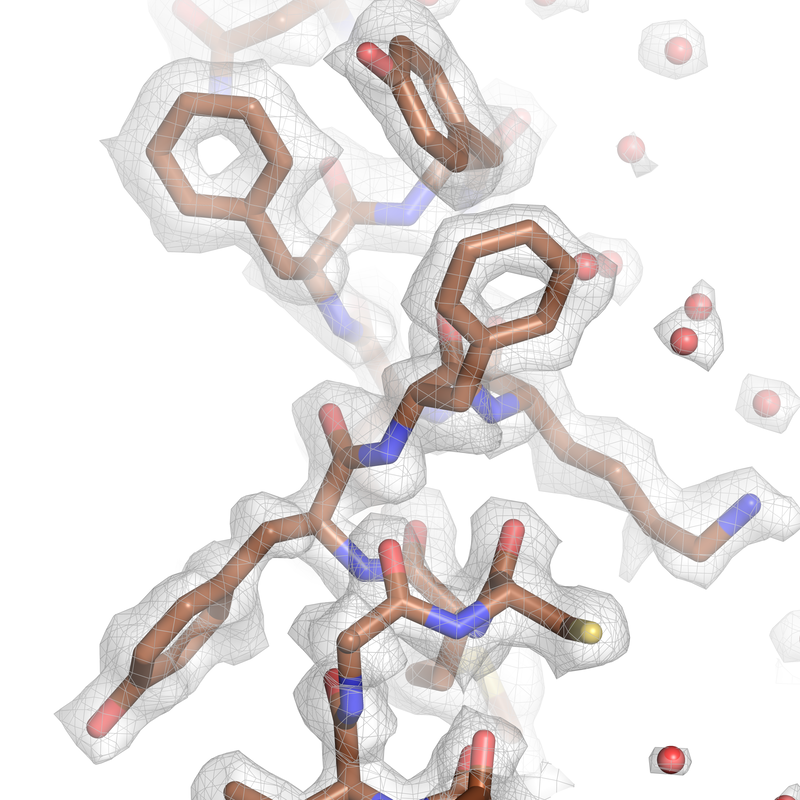
Cryo-EM map of the human CDK-activating kinase at 2 Å resolution. This density provides detailed information about the protein structure and allows identification of bound water molecules and post-translational modifications.
Cushing VI, Koh AF, Feng J, Jurgaityte K, Bondke A, Kroll SHB, Barbazanges M, Scheiper B, Bahl AK, Barret AGM, Ali S#, Kotecha A#, Greber BJ# (2024). High-resolution cryo-electron microscopy of the human CDK-activating kinase for structure-based drug design. Nat. Commun. 15: 2265. #corresponding authors Link bioRxiv, doi: 2023.04.07.536029. #corresponding authors Link Greber BJ# (2024) High-resolution cryo-EM of a small protein complex: The structure of the human CDK-activating kinase. Structure 32: 380-392, doi: 10.1016/j.str.2024.03.003. #corresponding author Link |